
INTRODUCTION TO GENOMICS
SECOND EDITION[1]
Author: Arthur M. Lesk
Publisher: Oxford University Press Inc., 2012, New York, 420 pages
Reviewed by:
Dr Ahmed Osman
Professor Abdul-Badi Abou-Samra
Relevant topics to the project of Genomics were carefully selected and summarized in a simplified way to be easily understood by non-medical professionals.
The Human Genome
Genes are composed of double helical deoxyribonucleic acids, or DNA. DNA is formed from the repetition of 4 nucleotides, where each nucleotide is formed from a nitrogen base (adenine, cytosine, thymine and guanine), a deoxyribose molecule and phosphoric acid; thus 4 different nucleotides exist in DNA. The nucleotides are named after the nitrogen base that exist in them and thus are abbreviated as A, T, G and C. The double stranded DNA molecules are formed by the pairing of 2 nucleotides with each other; C pairs with G and A pairs with T. During cell division, the double helix DNA open up and each DNA strand becomes a template for the formation of new double helical DNA that is identical to the original one. This way, the genetic information is passed from one “mother” cell to its 2 “daughter” cells.
The human genome contains approximately 3.2 billion base pairs, distributed among 22 paired chromosomes, plus two X chromosomes in females and one X and one Y chromosome in males. The first human genomes were determined in 2001, the culmination of 10 years of pioneering work and dedication. Since then, advances in technology have made genomic sequencing cheaper and faster. Sequence data now flow copiously, creating the challenges of understanding the information that our genomes contain, and applying the data and analysis to improve human welfare. Sequencing genomes of other species facilitates and extends these goals by revealing general principles of biology.
How do the contents of our genomes determine who we are?
Who we are is defined by our phenotype, which is the collection of all of our observable traits. These include macroscopic properties such as height, weight, eye and hair, and microscopic ones, such as sickle-cell anemia. The phenotype is determined by our genes, environment, life history and epigenetics. While our genes are encoded by a varying sequence of nucleotides, the genotype is determined by the DNA sequence, both nuclear and mitochondrial. Epigenetics refer to how the environment and our life history affect our genes and modify them chemically to influence their expression.
Traits that govern the susceptibility to disease and risk factors are of great importance to clinical applications of genomics and to those that determine the effectiveness of different drugs in different individuals. These allow for personalized prevention and treatment of disease based on DNA sequences, or pharmacogenomics.
Our life history includes the total integration of our experiences, and the physical and psychological environment in which we have developed, while the nutritional history influences physical development. Both nurturing environments and educational opportunities influence our mental development. The growing recognition of the importance of in utero environment in determining the developmental curve, and even adult characteristics, is obscure than most aspects of our life history.
The relative importance of these factors in determining our phenotype varies from trait to trait. Some traits are determined solely and irrevocably by our alleles for specific genes. Others, on the other hand, depend on complex interactions between genes and life history and epigenetic signals from parents.
Consequently, phenotypic traits – both macroscopic and molecular – depend on a combination of influences from genome sequences, the individual’s life history and the epigenetic signals in the fertilized egg.
Contents of the Human Genome
The most prominent and familiar aspects of the genome are the regions that code for proteins. Some regions of the genome encode non-protein-coding RNA like transfer RNAs, the RNA components of ribosomes, microRNAs and small interfering RNAs that regulate translation. Other regions contain binding sites for ligands responsible for the regulation of transcription. Repetitive elements of an unknown function surprisingly account for the large fractions of our genomes. Long and Short Interspersed Elements (LINES and SINES) account for 21% and 13% of the genome, respectively. Even more-highly repeated sequences – minisatellites, microsatellites and telomeres – may appear as tens or even hundreds of thousands of copies, in aggregate amounting to 15% of the genome.
Protein Synthesis
Transcription of a protein-coding gene into RNA is followed in eukaryotes by splicing to remove non-coding sequences and to form a mature messenger RNA (mRNA) molecule. Most human protein-coding genes contain exons (expressed regions) interrupted by introns (regions spliced out of mRNA and not translated to protein). The ribosome synthesizes a polypeptide chain according to the sequence of triplets of nucleotides, or codons, in the mRNA. The protein folds spontaneously into a native three-dimensional structure that accounts for its biological function. In short, DNA makes the RNA that makes Protein. The codons are those appearing in DNA rather than RNA; that is, the codons contain T rather than U. Multiple codons specify the same amino acid, e.g. GTT, GTC and GTA code for valine. Three triplets – TAA, TIA and TAG – function as STOP signals, affecting the termination of translation. Alternative splicing involves forming a mature messenger RNA from different choices of exons from a gene, but always in the order in which they appear in the genome. RNA editing involves changing codons and adenine to inosine, for example. So, alternative splicing and editing result in different proteins that are tissue-specific. Pseudogenes, though, are degenerate genes that have mutated so far from their original sequences that the polypeptide sequence they encode will not be functional.
Genome Sequencing
This is the process of determining the complete DNA sequence of an organism's genome at a single time. This entails sequencing all of an organism's chromosomal DNA as well as DNA contained in the mitochondria and, for plants, in the chloroplast. Since many different species have now had their genomes sequenced, this helps us to understand the functions of different regions of the human genome. Many people have undergone genetic testing: for example, many prospective parents determine whether they are carriers of cystic fibrosis or not. Many women test for potentially dangerous mutations in genes that can predispose an individual to cancer, such as BRCA1 and BRCA2 (cancer-related genes). Law-enforcement agencies determine DNA sequences from samples from crime scenes. A separate area of comparative genomics involves comparing the genome sequences of normal and cancer cells from patients. These can differ in essential ways, which assist in the precise diagnosis and guide treatment.
Resequencing and exome sequencing is the process of mapping onto the reference genome. This helps determine the variation in the genome of an individual from the reference genome. By correlating these variations with phenotype – for example, the presence of an inherited disease – it is possible to identify the genetic origin of the lesion. Many inherited diseases result from loss of activity of particular proteins. The loss of activity frequently arises from a specific mutation in the sequence coding for the protein. To identify such a mutation, it is not necessary to sequence the entire genome, but only the protein-coding regions; namely, the exons.
Variations within and between populations
Any two people, except for identical siblings, have genomic sequences that differ at approximately 0.1% of the positions. Measurements of multiple human genomes permit distinction between random components of this variation and those that systematically characterize different populations. Many, but not all, of the variation takes the form of isolated base substitutions, or single-nucleotide polymorphisms.
Sequence variation in humans has applications in anthropology to trace migration patterns, and in personal identification to prove paternity or for crime investigation. Several companies now offer personal genome sequencing. Many provide sequencing of mitochondrial DNA or individual loci in nuclear DNA, for the tracing of ancestry. The application of DNA to demonstrate legal relationships is the most common, where paternity testing is well established.
The human genome and medicine
Understanding individual genetic predispositions to disease can, for some conditions, help preserve health through suggesting changes in lifestyle and/or medical treatment. An example of a risk factor detectable at the genetic level involves α1-antitrypsin, a protein that normally functions to inhibit elastase in the alveoli of the lung. People homozygous for the Z mutant of α1-antitrypsin (342Glu→Lys) only express a dysfunctional protein. They are at risk of emphysema because of damage to the lungs from endogenous elastase unchecked by normal inhibitory activity, and also of liver disease because of the accumulation of a polymeric form of α1-antitrypsin in liver cells where it is synthesized. Smoking makes the development of emphysema all but certain. Heavy smokers homozygous for Z-antitrypsin generally die from respiratory disease by the age of 50. In these cases, the disease is brought on by a combination of genetic and environmental factors. In other cases, detection of genetic abnormalities will not prevent a disease but can dispel fear of the unknown; Huntington disease is a good example, which is an inherited rare neurodegenerative disorder. Its symptoms are quite severe, including uncontrollable dance-like movements, mental disturbance, personality changes and intellectual impairment. Death usually follows within 10–15 years of the onset of symptoms. Genetic counseling is used as a potential preventative approach in avoiding abnormalities or diseases that arise from dangerous combinations of parental genes.
Early detection of many diseases permits simpler and more successful treatment. For instance, oncologists classify leukemia (blood cancer) into seven subtypes. Determination of the subtype from gene expression patterns permits better prognosis and treatment.
Recent scientific advances have accelerated the drug development process. Identifying metabolic features unique to a pathogen helps to identify targets for antibacterial and antiviral agents. Human proteins provide other drug targets to deal with molecular dysfunction or to adjust regulatory controls. Knowing the structure of a target permits computer-assisted drug design by molecular modeling.
Genomic screening has a great impact on healthcare delivery. For example, many drugs vary in their effectiveness in different patients. A drug may be effective for some patients and useless for others. Some patients may tolerate a treatment easily; others may suffer side effects ranging from discomfort, through disability, to death. Analysis of patients’ genes and proteins permits selection of drugs and dosages optimal for individual patients; a field called pharmacogenomics. Physicians can thereby avoid experimenting with different therapies, a procedure that is dangerous in terms of side effects, sometimes even fatal, time-consuming and expensive. Treatment of patients for adverse reactions to prescribed drugs consumes billions of dollars in healthcare costs. A good example here is Abacavir, which is a drug used in treatment of AIDS. 4–8% of patients show a serious, potentially fatal, hypersensitivity reaction. This is correlated with MHC allele HLA-B*5701. Genomic screening can thereby detect potential hypersensitivity, and guide treatment.
Genetic diseases – some examples of their causes and treatment
Mutation: This is the permanent alteration of the nucleotide sequence of the genome of an organism, virus, or extra chromosomal or other genetic elements. Mutations result from errors during DNA replication or other types of damage to DNA, which may then undergo error-prone repair or cause an error during other forms of repair. Mutations may also result from insertion or deletion of segments of DNA due to mobile genetic elements. Mutations may or may not produce discernible changes in the observable characteristics (phenotype) of an organism (silent mutation). Mutations play a part in both normal and abnormal biological processes including evolution, cancer, and the development of the immune system:
- Missense mutations lead to a change in one or more amino acids in the encoded protein – sickle-cell mutation is an example of a missense mutation;
- Nonsense mutations lead to an early stop signal and a shorter protein;
- Mutations in regulatory regions of a gene change the expression of the protein;
- Mutations in splice sites lead to pre-mature mRNA which can be translated into a different protein or a shorter protein;
- Deletion and insertion.
Sickle-cell anemia: Patterns of inheritance showed that sickle-cell anemia is a genetic disease. The sickle-cell mutation changes residue 6 of the b chain from a charged side chain, glutamic acid, to a non-polar one, valine (b6Glu→Val). This mutant protein precipitates in the red blood cells and form sickle-like shape, and this in turn blocks the capillaries and leads to decrease oxygen delivery to tissues, red cells destruction, anemia and jaundice.
Phenylketonuria (PKU) is a genetic disease caused by deficiency in a metabolic enzyme, phenylalanine hydroxylase; the enzyme that converts phenylalanine to tyrosine. If untreated, phenylalanine accumulates in the blood to toxic levels, leading to high levels of phenylalanine that cause a variety of developmental defects, including mental retardation, microcephaly and seizures. The disease cannot be cured, but symptoms can be avoided by lifestyle control: a phenylalanine-free diet. Screening of newborns for PKU is legally required in the United States and many other countries. PKU is an example of how understanding the molecular mechanism of a disease can, for some conditions, help restore and preserve health through suggested changes in lifestyle and/or medical treatment.
Genetics and cancer: The onset of cancer is associated with loss of genome integrity. Cancer results from accumulated mutations that break down the controls on cell growth. The source can be in three classes of genes: genes that regulate cell proliferation; genes required for repair of DNA damage; and genes that control apoptosis (programmed cell death). The ‘two-hit hypothesis’ interprets the relation between sporadic and familial forms of the disease that mutate both copies of such genes, such as retinoblastoma – a rare childhood tumor of the eye. Approximately 30–40% of cases are familial; the rest are sporadic. The familial form shows an autosomal dominant inheritance pattern. The idea is that non-familial cases require inactivation of both copies of the retinoblastoma gene, each of which was originally functional, while separate and independent mutations would be necessary. In contrast, familial retinoblastoma affects a person who has inherited one defective and one functional copy of the gene. That is, the first hit is inherited; all that is needed is the second hit.
The Philadelphia chromosome is an abnormal, shortened chromosome 22, arising from a translocation, an exchange of chromosomal segments between chromosome 22 and chromosome 9. This translocation leads to formation of a protein insensitive to normal regulation, and hence the development of leukemia. The discovery of this lead to a revolution in the treatment of leukemia by imatinib mesylate.
Genomics in personal identification
Legal applications of DNA sequencing depend on several scientific facts:
- The genomes of all individuals, except identical siblings, are unique. Like fingerprints, genomes provide a unique personal identification. A blood stain at a crime scene, like a set of fingerprints, can be traced to a specific individual.
- The genome of every person combines chromosomes from his or her parents. Therefore, unlike fingerprints, genomes can indicate familial relationships; notably, identification of paternity.
- Each person’s genome contains genes that influence, even if they do not inevitably determine, recognizable features, such as eye color. In principle, DNA left by an unknown individual at a crime scene could be analyzed to suggest a physical description of the source individual.
- Thus, unlike fingerprints, genomes contain surplus information about a person than simple identification. The treatment of this information by governmental authorities raises ethical and legal questions.
The use of molecular characteristics to identify people and relationships is a century old. The earliest methods applied the classical blood groups – A, B, AB, and O. A suspect with blood type O must be innocent of a crime committed by a person of blood type A. A person of blood type O could not be the parent of a child of type AB. However, many people share the same blood type. Therefore, blood typing can prove innocence but not guilt. In contrast, DNA sequences can provide positive proof of guilt or paternity.
Mitochondrial DNA
Human mitochondrial DNA contains a hyper variable 100 bp region, which varies by 1–2% between unrelated individuals. The mitochondrial DNA of unrelated people typically differs at eight positions.
DNA microarray (also commonly known as DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. The transcriptome of a cell is the set of RNA molecules it contains; the proteome is its proteins. Expression patterns can help identify genes that underlie diseases. Some diseases, such as cystic fibrosis, arise from mutations in single genes. For these, isolating a region by genetic mapping can help pinpoint the lesion. Other diseases, such as asthma, depend on interactions among many genes and interaction with many environmental factors. Understanding the etiology of multi-factorial diseases requires the ability to determine and analyze expression patterns of many genes, which may be distributed around different chromosomes.
Examples of DNA microarrays applications:
- Comparison of related species.
- Diagnosis of genetic disease.
- Precise diagnosis of disease. Different related types of leukemia can be distinguished by signature patterns of gene expression. Knowing the exact type of the disease is important for prognosis and for selecting optimal treatment.
- Drug selection and target selection for drug design.
- Determination of gene function.
Proteomics: The proteome is the entire set of proteins expressed by a genome, cell, tissue, or organism at a certain time.
Pattern of inheritance:
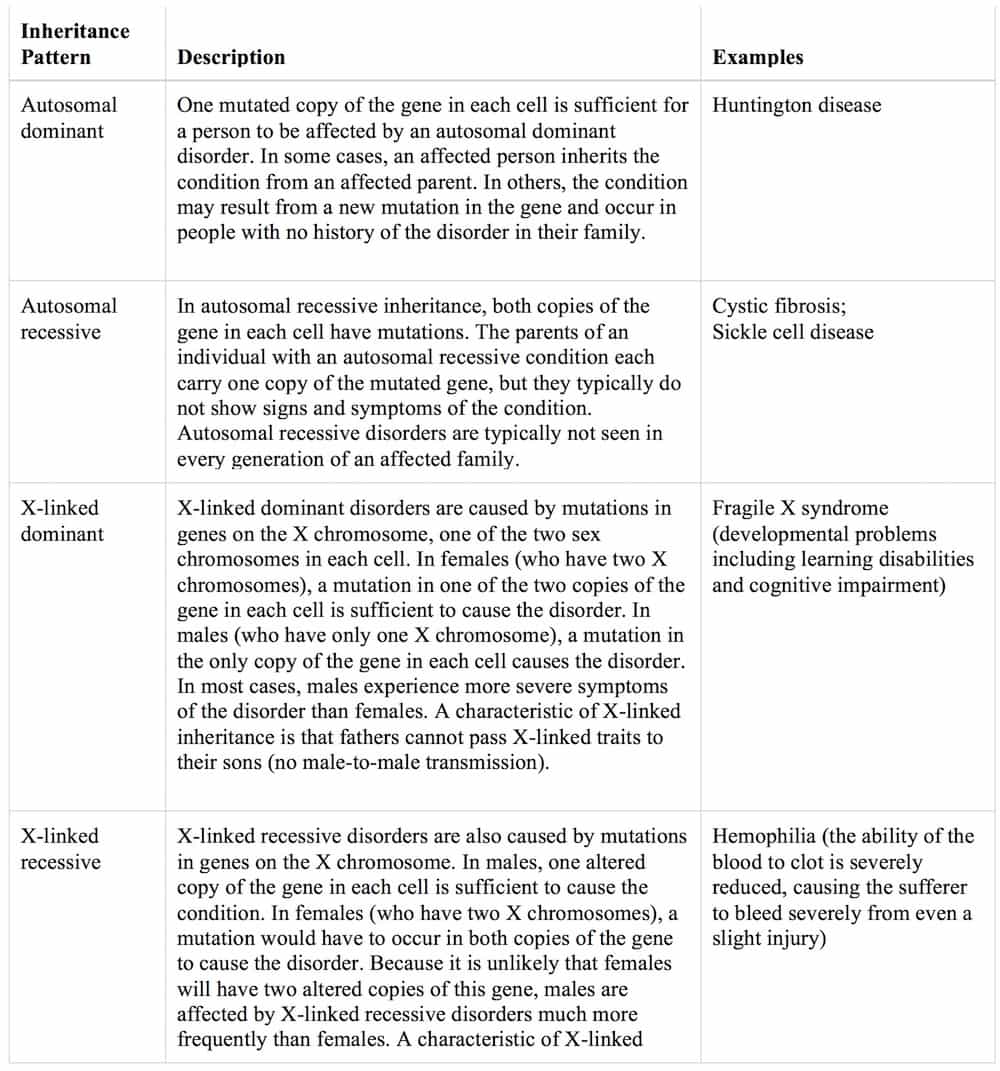
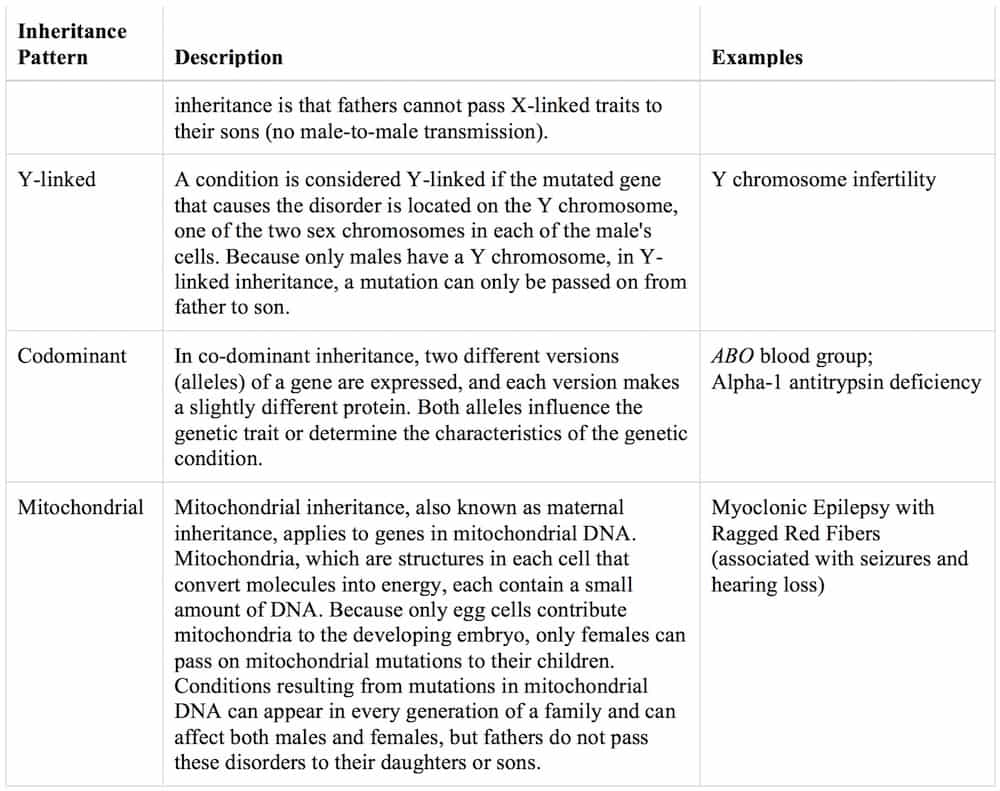
Disorders caused by changes in the number or structure of chromosomes also do not follow the straightforward patterns of inheritance listed above.
Examples:
Klinefelter syndrome: This occurs when a boy is born with one or more extra X chromosomes. Most males have one Y and one X chromosome. Having an extra X chromosomes can cause a male to have some physical traits unusual for males.
Down syndrome: Which is also known as trisomy 21, is a genetic disorder caused by the presence of all or part of a third copy of chromosome 21. It is typically associated with physical growth delays, characteristic facial features and mild to moderate intellectual disability. The average IQ of a young adult with Down syndrome is 50, equivalent to the mental ability of an 8- or 9-year-old child, but this can vary widely, as well as an increased risk of cancer.
[1] "This research was made possible by the NPRP grant “Indigenizing Genomics in the Gulf Region (IGGR): The Missing Islamic Bioethical Discourse", no. NPRP8-1620-6-057 from the Qatar National Research Fund (a member of The Qatar Foundation). The statements made herein are solely the responsibility of the author[s]."




















Add new comment